Interactive Visualizion with Open Altimetry & Google Earth Engine
Contents
Interactive Visualizion with Open Altimetry & Google Earth Engine#
By Philipp Arndt
Scripps Institution of Oceanography, University of California San Diego
Github: @fliphilipp
Contact: parndt@ucsd.edu
Learning Objectives#
Discover some interesting ICESat-2 data by browsing OpenAltimetry
Get the data into a Jupyter Notebook, plot it and overlay the ground track on a map
Use Google Earth Engine to find the closest-in-time Sentinel-2 image that is cloud-free along ICESat-2’s gound track
Make a plot from ICESat-2 data and contemporaneous imagery in Jupyter/matplotlib
Bonus Code: Use Google Earth Engine to extract Sentinel-2 data along the ICESat-2 ground track
Computing environment#
We’ll be using the following Python libraries in this notebook:
%matplotlib widget
import os
import ee
import geemap
import requests
import numpy as np
import pandas as pd
import matplotlib.pylab as plt
from datetime import datetime
from datetime import timedelta
from ipywidgets import Layout
import rasterio as rio
from rasterio import plot as rioplot
from rasterio import warp
The import below is a class that I wrote myself. It helps us read and store data from the OpenAltimetry API.
If you are interested in how this works, you can find the code in utils/oa.py.
from utils.oa import dataCollector
Google Earth Engine Authentication and Initialization#
GEE requires you to authenticate your access, so if ee.Initialize() does not work you first need to run ee.Authenticate(). This gives you a link at which you can use your google account that is associated with GEE to get an authorization code. Copy the authorization code into the input field and hit enter to complete authentication.
try:
ee.Initialize()
except:
ee.Authenticate()
ee.Initialize()
# ee.Authenticate()
Downloading data from the OpenAltimetry API#
Let’s say we have found some data that looks weird to us, and we don’t know what’s going on.
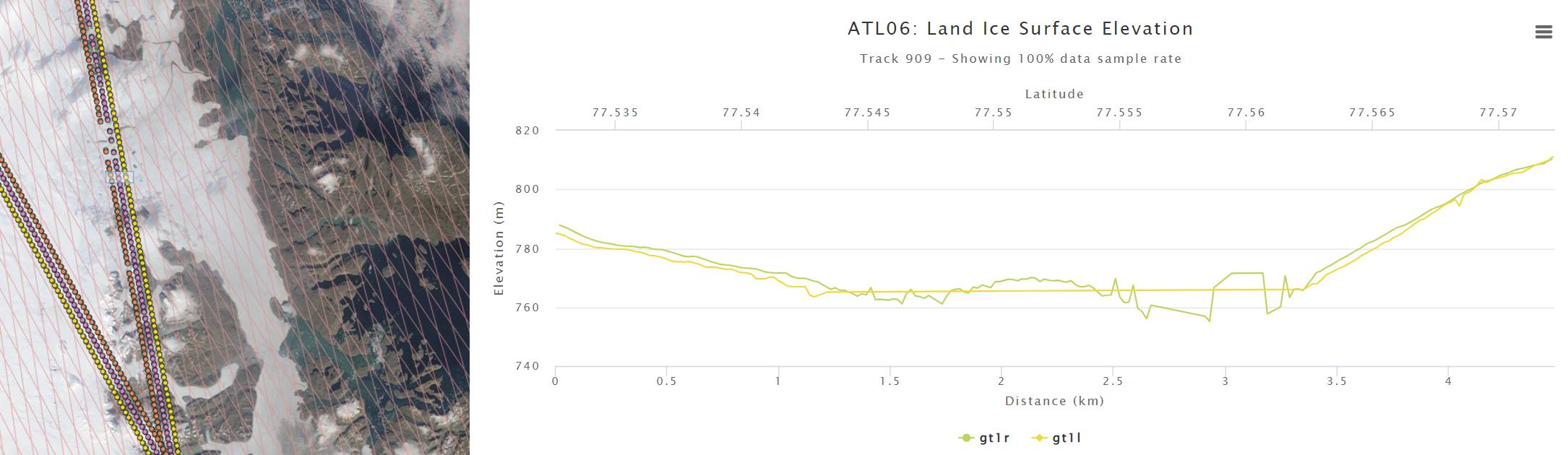
A short explanation of how I got the data:#
I went to openaltimetry.org and selected BROWSE ICESAT-2 DATA. Then I selected ATL 06 (Land Ice Height) on the top right, and switched the projection🌎 to Arctic. Then I selected August 22, 2021 in the calendar📅 on the bottom left, and toggled the cloud☁️ button to show MODIS imagery of that date. I then zoomed in on my region of interest.
To find out what ICESat-2 has to offer here, I clicked on SELECT A REGION on the top left, and drew a rectangle around that mysterious cloud. When right-clicking on that rectangle, I could select View Elevation profile. This opened a new tab, and showed me ATL06 and ATL08 elevations.
It looks like ATL06 can’t decide where the surface is, and ATL08 tells me that there’s a forest canopy on the Greenland Ice Sheet? To get to the bottom of this, I scrolled all the way down and selected 🛈Get API URL. The website confirms that the “API endpoint was copied to clipboard.” Now I can use this to access the data myself.
Note: Instead of trying to find this region yourself, you can access the OpenAltimetry page shown above by going to this annotation🏷️. Just left-click on the red box and select “View Elevation Profile”.
Getting the OpenAltimetry info into python#
All we need to do is to paste the API URL that we copied from the webpage into a string. We also need to specify which beam we would like to look at. The GT1R ground track looks funky, so let’s look at that one!
# paste the API URL from OpenAltimetry below, and specify the beam you are interested in
oa_api_url = 'http://openaltimetry.org/data/api/icesat2/atl03?date=2021-08-22&minx=-24.03772653601395&miny=77.53753839308544&maxx=-23.86438315078483&maxy=77.57604313591165&trackId=909&beamName=gt1r&beamName=gt1l&outputFormat=json'
gtx = 'gt1r'
We can now initialize a dataCollector object, using the copy-pasted OpenAltimetry API URL, and the beam we would like to look at.
(Again, I defined this class in utils/oa.py to do some work for us in the background.)
is2data = dataCollector(oaurl=oa_api_url,beam=gtx, verbose=True)
OpenAltimetry API URL: http://openaltimetry.org/data/api/icesat2/atlXX?date=2021-08-22&minx=-24.03772653601395&miny=77.53753839308544&maxx=-23.86438315078483&maxy=77.57604313591165&trackId=909&outputFormat=json&beamName=gt1r&client=jupyter
Date: 2021-08-22
Track: 909
Beam: gt1r
Latitude limits: [77.53753839308544, 77.57604313591165]
Longitude limits: [-24.03772653601395, -23.86438315078483]
Alternatively, we could use a date, track number, beam, and lat/lon bounding box as input to the dataCollector.
date = '2021-08-22'
rgt = 909
beam = 'gt1r'
latlims = [77.5326, 77.5722]
lonlims = [-23.9891, -23.9503]
is2data = dataCollector(date=date, latlims=latlims, lonlims=lonlims, track=rgt, beam=gtx, verbose=True)
OpenAltimetry API URL: https://openaltimetry.org/data/api/icesat2/atlXX?date=2021-08-22&minx=-23.9891&miny=77.5326&maxx=-23.9503&maxy=77.5722&trackId=909&beamName=gt1r&outputFormat=json&client=jupyter
Date: 2021-08-22
Track: 909
Beam: gt1r
Latitude limits: [77.5326, 77.5722]
Longitude limits: [-23.9891, -23.9503]
Note that this also constructs the API url for us.
Requesting the data from the OpenAltimetry API#
Here we use the requestData() function of the dataCollector class, which is defined in utils/oa.py. It downloads all data products that are available on OpenAltimetry based on the inputs with which we initialized our dataCollector, and writes them to pandas dataframes.
is2data.requestData(verbose=True)
---> requesting ATL03 data... 6314 data points.
---> requesting ATL06 data... 198 data points.
---> requesting ATL07 data... No data.
---> requesting ATL08 data... 26 data points.
---> requesting ATL10 data... No data.
---> requesting ATL12 data... No data.
---> requesting ATL13 data... No data.
The data are now stored as data frames in our dataCollector object. To verify this, we can run the cell below.
vars(is2data)
{'url': 'https://openaltimetry.org/data/api/icesat2/atlXX?date=2021-08-22&minx=-23.9891&miny=77.5326&maxx=-23.9503&maxy=77.5722&trackId=909&beamName=gt1r&outputFormat=json&client=jupyter',
'date': '2021-08-22',
'track': 909,
'beam': 'gt1r',
'latlims': [77.5326, 77.5722],
'lonlims': [-23.9891, -23.9503],
'atl03': lat lon h conf
0 77.572273 -23.953863 748.45350 Noise
1 77.572246 -23.953913 847.64276 Noise
2 77.572247 -23.953895 785.39874 Noise
3 77.572232 -23.953940 906.48040 Noise
4 77.572223 -23.953903 739.40704 Noise
... ... ... ... ...
6309 77.532606 -23.986983 788.30840 High
6310 77.532600 -23.986988 787.72420 High
6311 77.532594 -23.986993 788.89220 High
6312 77.532594 -23.986993 786.84937 High
6313 77.532594 -23.986993 787.69586 High
[6314 rows x 4 columns],
'atl06': lat lon h
0 77.572104 -23.954022 810.27400
1 77.571928 -23.954170 809.49520
2 77.571751 -23.954318 808.62430
3 77.571575 -23.954465 808.51855
4 77.571398 -23.954612 808.20930
.. ... ... ...
193 77.533473 -23.986256 785.09015
194 77.533297 -23.986406 785.85700
195 77.533120 -23.986554 786.59470
196 77.532944 -23.986702 787.29694
197 77.532768 -23.986849 787.82263
[198 rows x 3 columns],
'atl07': Empty DataFrame
Columns: [lat, lon, h]
Index: [],
'atl08': lat lon h canopy
0 77.571838 -23.954245 809.23700 3.455017
1 77.570961 -23.954979 807.26886 NaN
2 77.570076 -23.955711 804.86220 NaN
3 77.569191 -23.956451 801.61490 NaN
4 77.568314 -23.957195 797.30896 NaN
5 77.567429 -23.957932 792.91420 2.570374
6 77.566544 -23.958658 788.39480 1.922424
7 77.565666 -23.959396 784.11694 2.658264
8 77.564781 -23.960142 779.79500 3.664246
9 77.563904 -23.960869 776.76560 4.249573
10 77.563019 -23.961609 767.44324 6.668396
11 77.562141 -23.962358 762.40380 10.477356
12 77.544502 -23.977083 763.87840 8.553955
13 77.543617 -23.977800 764.07556 7.998474
14 77.542732 -23.978540 765.19090 6.761414
15 77.541855 -23.979282 768.75446 4.206970
16 77.540970 -23.980009 771.93854 NaN
17 77.540085 -23.980747 773.26587 NaN
18 77.539207 -23.981489 774.43463 NaN
19 77.538322 -23.982227 776.89930 NaN
20 77.537445 -23.982954 777.96080 NaN
21 77.536560 -23.983694 779.56340 NaN
22 77.535675 -23.984425 769.83990 11.356079
23 77.534798 -23.985142 770.10900 11.810181
24 77.533913 -23.985880 772.83750 11.275330
25 77.533035 -23.986628 775.42786 12.219727,
'atl10': Empty DataFrame
Columns: [lat, lon, h]
Index: [],
'atl12': Empty DataFrame
Columns: [lat, lon, h]
Index: [],
'atl13': Empty DataFrame
Columns: [lat, lon, h]
Index: []}
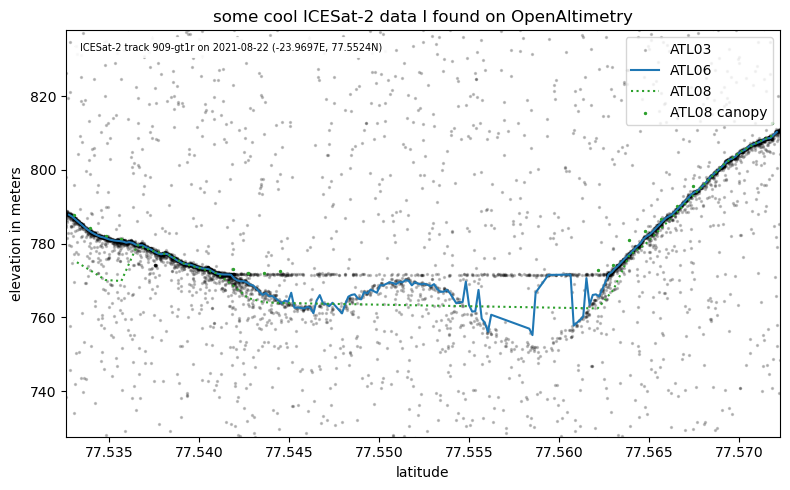
Plotting the ICESat-2 data#
Now let’s plot this data. Here, we are just creating an empty figure fig with axes ax.
# create the figure and axis
fig, ax = plt.subplots(figsize=[9,5])
# plot the data products
atl06, = ax.plot(is2data.atl06.lat, is2data.atl06.h, c='C0', linestyle='-', label='ATL06')
atl08, = ax.plot(is2data.atl08.lat, is2data.atl08.h, c='C2', linestyle=':', label='ATL08')
if np.sum(~np.isnan(is2data.atl08.canopy))>0:
atl08canopy = ax.scatter(is2data.atl08.lat, is2data.atl08.h+is2data.atl08.canopy, s=2, c='C2', label='ATL08 canopy')
# add labels, title and legend
ax.set_xlabel('latitude')
ax.set_ylabel('elevation in meters')
ax.set_title('Some ICESat-2 data I found on OpenAltimetry!')
ax.legend(loc='upper left')
# add some text to provide info on what is plotted
info = 'ICESat-2 track {track:d}-{beam:s} on {date:s}\n({lon:.4f}E, {lat:.4f}N)'.format(track=is2data.track,
beam=is2data.beam.upper(),
date=is2data.date,
lon=np.mean(is2data.lonlims),
lat=np.mean(is2data.latlims))
infotext = ax.text(0.01, -0.08, info,
horizontalalignment='left',
verticalalignment='top',
transform=ax.transAxes,
fontsize=7,
bbox=dict(edgecolor=None, facecolor='white', alpha=0.9, linewidth=0))
# set the axis limits
ax.set_xlim((is2data.atl03.lat.min(), is2data.atl03.lat.max()))
ax.set_ylim((741, 818));
Let’s add the ATL03 photons to better understand what might be going on here.
atl03 = ax.scatter(is2data.atl03.lat, is2data.atl03.h, s=1, color='black', label='ATL03', zorder=-1)
ax.legend(loc='upper left')
fig.tight_layout()
Saving the plot to a file#
fig.savefig('my-plot.jpg', dpi=300)
To make plots easier to produce, the dataCollector class also has a method to plot the data that we downloaded.
fig = is2data.plotData();
fig
Ground Track Stats#
So far we have only seen the data in elevation vs. latitude space. It’s probably good to know what the scale on the x-axis is here in units that we’re familiar with.
def dist_latlon2meters(lat1, lon1, lat2, lon2):
# returns the distance between two coordinate points - (lon1, lat1) and (lon2, lat2) along the earth's surface in meters.
R = 6371000
def deg2rad(deg):
return deg * (np.pi/180)
dlat = deg2rad(lat2-lat1)
dlon = deg2rad(lon2-lon1)
a = np.sin(dlat/2) * np.sin(dlat/2) + np.cos(deg2rad(lat1)) * np.cos(deg2rad(lat2)) * np.sin(dlon/2) * np.sin(dlon/2)
c = 2 * np.arctan2(np.sqrt(a), np.sqrt(1-a))
return R * c
lat1, lat2 = is2data.atl08.lat[0], is2data.atl08.lat.iloc[-1]
lon1, lon2 = is2data.atl08.lon[0], is2data.atl08.lon.iloc[-1]
ground_track_length = dist_latlon2meters(lat1, lon1, lat2, lon2)
print('The ground track is about %.1f kilometers long.' % (ground_track_length/1e3))
The ground track is about 4.4 kilometers long.
Google Earth Engine#
Google Earth Engine (GEE) has a large catalog of geospatial raster data, which is ready for analysis in the cloud. It also comes with an online JavaScript code editor.
 But since we all seem to be using python, it would be nice to have these capabilities available in our Jupyter comfort zone…
But since we all seem to be using python, it would be nice to have these capabilities available in our Jupyter comfort zone…
Thankfully, there is a python API for GEE, which we have imported using import ee earlier. It doesn’t come with an interactive map, but the python package geemap has us covered!
Show a ground track on a map#
We can start working on our map by calling geemap.Map(). This just gives us a world map with a standard basemap.
from ipywidgets import Layout
Map = geemap.Map(layout=Layout(width='70%', max_height='450px'))
Map
Now we need to add our ICESat-2 gound track to that map. Let’s use the lon/lat coordinates of the ATL08 data product for this.
We also need to specify which Coordinate Reference System (CRS) our data is in. The longitude/latitude system that we are all quite familiar with is referenced by EPSG:4326. To add the ground track to the map we need to turn it into an Earth Engine “Feature Collection”.
ground_track_coordinates = list(zip(is2data.atl08.lon, is2data.atl08.lat))
ground_track_projection = 'EPSG:4326' # <-- this specifies that our data longitude/latitude in degrees [https://epsg.io/4326]
gtx_feature = ee.FeatureCollection(ee.Geometry.LineString(coords=ground_track_coordinates,
proj=ground_track_projection,
geodesic=True))
gtx_feature
- type:FeatureCollection
- system:index:String
- type:Feature
- id:0
- type:LineString
- 0:-23.95424461364746
- 1:77.57183837890625
- 0:-23.954978942871094
- 1:77.57096099853516
- 0:-23.955711364746094
- 1:77.57007598876953
- 0:-23.956451416015625
- 1:77.5691909790039
- 0:-23.957195281982422
- 1:77.56831359863281
- 0:-23.957931518554688
- 1:77.56742858886719
- 0:-23.95865821838379
- 1:77.56654357910156
- 0:-23.959396362304688
- 1:77.56566619873047
- 0:-23.960142135620117
- 1:77.56478118896484
- 0:-23.96086883544922
- 1:77.56390380859375
- 0:-23.96160888671875
- 1:77.56301879882812
- 0:-23.962358474731445
- 1:77.56214141845703
- 0:-23.977083206176758
- 1:77.54450225830078
- 0:-23.977800369262695
- 1:77.54361724853516
- 0:-23.978540420532227
- 1:77.54273223876953
- 0:-23.97928237915039
- 1:77.54185485839844
- 0:-23.980009078979492
- 1:77.54096984863281
- 0:-23.98074722290039
- 1:77.54008483886719
- 0:-23.981489181518555
- 1:77.5392074584961
- 0:-23.982227325439453
- 1:77.53832244873047
- 0:-23.982954025268555
- 1:77.53744506835938
- 0:-23.983694076538086
- 1:77.53656005859375
- 0:-23.984424591064453
- 1:77.53567504882812
- 0:-23.98514175415039
- 1:77.53479766845703
- 0:-23.98587989807129
- 1:77.5339126586914
- 0:-23.98662757873535
- 1:77.53303527832031
Now that we have it in the right format, we can add it as a layer to the map.
Map.addLayer(gtx_feature, {'color': 'red'}, 'ground track')
According to the cell above this should be a red line. But we still can’t see it, because we first need to tell the map where to look for it.
Let’s calculate the center longitude and latitude, and center the map on it.
center_lon = (lon1 + lon2) / 2
center_lat = (lat1 + lat2) / 2
Map.setCenter(center_lon, center_lat, zoom=4);
So we actually couldn’t see it because it was in Greenland.
Unfortunately the basemap here doesn’t give us much more information. Let’s add a satellite imagery basemap.
This is a good time to look at the layer control on the top right.
Map.add_basemap('SATELLITE') # <-- this adds a layer called 'Google Satellite'
Map.layer_opacity(name='Google Satellite', opacity=0.5)
Map.setCenter(center_lon, center_lat, zoom=7);
Map.addLayer(gtx_feature,{'color': 'red'},'ground track')
…looks like this basemap still doesn’t give us any more clues about the nature of this weird ICESat-2 data. Let’s dig deeper.
Query for Sentinel-2 images#
Both of these Sentinel-2 satellites take images of most places on our planet at least every week or so. Maybe these images can tell us what was happening here around the same time that ICESat-2 acquired our data?
The imagery scenes live in image collections on Google Earth Engine.
You can find all collections here: https://developers.google.com/earth-engine/datasets/catalog/
The above link tells us we can find some images under 'COPERNICUS/S2_SR_HARMONIZED'.
collection_name1 = 'COPERNICUS/S2_SR_HARMONIZED' # Landsat 8 earth engine collection
# https://developers.google.com/earth-engine/datasets/catalog/LANDSAT_LC08_C01_T2
Access an image collection#
To access the collection, we call ee.ImageCollection:
collection = ee.ImageCollection(collection_name1)
collection
<ee.imagecollection.ImageCollection object at 0x7f8c097db9d0>
Can we find out how many images there are in total?
number_of_scenes = collection.size()
print(number_of_scenes)
ee.Number({
"functionInvocationValue": {
"functionName": "Collection.size",
"arguments": {
"collection": {
"functionInvocationValue": {
"functionName": "ImageCollection.load",
"arguments": {
"id": {
"constantValue": "COPERNICUS/S2_SR_HARMONIZED"
}
}
}
}
}
}
})
Actually, asking for the size of the collection does not do anything! 🤔
It just tells Earth Engine on the server-side that this variable refers to the size of the collection, which we may need later to do some analysis on the server. As long as this number is not needed, Earth Engine will not go through the trouble actually computing it.
To force Earth Engine to compute and get any information on the client side (our local machine / Cryocloud), we need to call .getInfo(). In this case that would be number_of_scenes = collection.size().getInfo().
Because this command would ask Earth Engine to count every single Sentinel-2 file that exists, this command would take a really long time to execute. I will avoid this here and just give you the answer from when I wrote this tutorial.
# number_of_scenes = collection.size().getInfo()
number_of_scenes = 19323842
print('There are %i number of scenes in the image collection' % number_of_scenes)
There are 19323842 number of scenes in the image collection
Filter an image collection by location and time#
Who wants to look at almost 20 million pictures? I don’t. So let’s try to narrow it down.
Let’s start with only images that overlap with the center of our ground track.
# the point of interest (center of the track) as an Earth Engine Geometry
point_of_interest = ee.Geometry.Point(center_lon, center_lat)
collection = collection.filterBounds(point_of_interest)
print('There are {number:d} images in the spatially filtered collection.'.format(number=collection.size().getInfo()))
There are 2782 images in the spatially filtered collection.
Much better! Now let’s only look at images that were taken soon before or after ICESat-2 passed over this spot.
days_buffer_imagery = 6
dateformat = '%Y-%m-%d'
datetime_requested = datetime.strptime(is2data.date, dateformat)
search_start = (datetime_requested - timedelta(days=days_buffer_imagery)).strftime(dateformat)
search_end = (datetime_requested + timedelta(days=days_buffer_imagery)).strftime(dateformat)
print('Search for imagery from {start:s} to {end:s}.'.format(start=search_start, end=search_end))
Search for imagery from 2021-08-16 to 2021-08-28.
collection = collection.filterDate(search_start, search_end)
print('There are {number:d} images in the spatially filtered collection.'.format(number=collection.size().getInfo()))
There are 34 images in the spatially filtered collection.
We can also sort the collection by date ('system:time_start'), to order the images by acquisition time.
collection = collection.sort('system:time_start')
Get image collection info#
Again, we need to use .getInfo() to actually see any information on our end. This is a python dictionary.
info = collection.getInfo()
type(info)
dict
Let’s see what’s inside!
info.keys()
dict_keys(['type', 'bands', 'id', 'version', 'properties', 'features'])
'features' sounds like it could hold information about the images we are trying to find…
len(info['features'])
34
A list of 34 things! Those are probably the 34 images in the collection. Let’s pick the first one and dig deeper!
feature_number = 0
info['features'][0].keys()
dict_keys(['type', 'bands', 'version', 'id', 'properties'])
info['features'][feature_number]['id']
'COPERNICUS/S2_SR_HARMONIZED/20210816T150759_20210816T150915_T27XVG'
Looks like we found a reference to a Sentinel-2 image! Let’s look at the 'bands'.
for band in info['features'][feature_number]['bands']:
print(band['id'], end=', ')
B1, B2, B3, B4, B5, B6, B7, B8, B8A, B9, B11, B12, AOT, WVP, SCL, TCI_R, TCI_G, TCI_B, MSK_CLDPRB, MSK_SNWPRB, QA10, QA20, QA60,
'properties' could be useful too!
info['features'][0]['properties'].keys()
dict_keys(['DATATAKE_IDENTIFIER', 'AOT_RETRIEVAL_ACCURACY', 'SPACECRAFT_NAME', 'SATURATED_DEFECTIVE_PIXEL_PERCENTAGE', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B8A', 'CLOUD_SHADOW_PERCENTAGE', 'MEAN_SOLAR_AZIMUTH_ANGLE', 'system:footprint', 'VEGETATION_PERCENTAGE', 'SOLAR_IRRADIANCE_B12', 'SOLAR_IRRADIANCE_B10', 'SENSOR_QUALITY', 'SOLAR_IRRADIANCE_B11', 'GENERATION_TIME', 'SOLAR_IRRADIANCE_B8A', 'FORMAT_CORRECTNESS', 'CLOUD_COVERAGE_ASSESSMENT', 'THIN_CIRRUS_PERCENTAGE', 'system:time_end', 'WATER_VAPOUR_RETRIEVAL_ACCURACY', 'system:time_start', 'DATASTRIP_ID', 'PROCESSING_BASELINE', 'SENSING_ORBIT_NUMBER', 'NODATA_PIXEL_PERCENTAGE', 'SENSING_ORBIT_DIRECTION', 'GENERAL_QUALITY', 'GRANULE_ID', 'REFLECTANCE_CONVERSION_CORRECTION', 'MEDIUM_PROBA_CLOUDS_PERCENTAGE', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B8', 'DATATAKE_TYPE', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B9', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B6', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B7', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B4', 'MEAN_INCIDENCE_ZENITH_ANGLE_B1', 'NOT_VEGETATED_PERCENTAGE', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B5', 'RADIOMETRIC_QUALITY', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B2', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B3', 'MEAN_INCIDENCE_ZENITH_ANGLE_B5', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B1', 'MEAN_INCIDENCE_ZENITH_ANGLE_B4', 'MEAN_INCIDENCE_ZENITH_ANGLE_B3', 'MEAN_INCIDENCE_ZENITH_ANGLE_B2', 'MEAN_INCIDENCE_ZENITH_ANGLE_B9', 'MEAN_INCIDENCE_ZENITH_ANGLE_B8', 'MEAN_INCIDENCE_ZENITH_ANGLE_B7', 'DARK_FEATURES_PERCENTAGE', 'HIGH_PROBA_CLOUDS_PERCENTAGE', 'MEAN_INCIDENCE_ZENITH_ANGLE_B6', 'UNCLASSIFIED_PERCENTAGE', 'MEAN_SOLAR_ZENITH_ANGLE', 'MEAN_INCIDENCE_ZENITH_ANGLE_B8A', 'RADIATIVE_TRANSFER_ACCURACY', 'MGRS_TILE', 'CLOUDY_PIXEL_PERCENTAGE', 'PRODUCT_ID', 'MEAN_INCIDENCE_ZENITH_ANGLE_B10', 'SOLAR_IRRADIANCE_B9', 'SNOW_ICE_PERCENTAGE', 'DEGRADED_MSI_DATA_PERCENTAGE', 'MEAN_INCIDENCE_ZENITH_ANGLE_B11', 'MEAN_INCIDENCE_ZENITH_ANGLE_B12', 'SOLAR_IRRADIANCE_B6', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B10', 'SOLAR_IRRADIANCE_B5', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B11', 'SOLAR_IRRADIANCE_B8', 'MEAN_INCIDENCE_AZIMUTH_ANGLE_B12', 'SOLAR_IRRADIANCE_B7', 'SOLAR_IRRADIANCE_B2', 'SOLAR_IRRADIANCE_B1', 'SOLAR_IRRADIANCE_B4', 'GEOMETRIC_QUALITY', 'SOLAR_IRRADIANCE_B3', 'system:asset_size', 'WATER_PERCENTAGE', 'system:index'])
That’s a lot going on right there! But 'GRANULE_ID' is probably useful. Let’s go through all our features and print the product id.
for feature in info['features']:
print(feature['properties']['GRANULE_ID'])
L2A_T27XVG_A023217_20210816T150915
L2A_T26XNM_A023217_20210816T150915
L2A_T27XVG_A023231_20210817T143747
L2A_T26XNM_A023231_20210817T143747
L2A_T27XVG_A032140_20210817T152909
L2A_T26XNM_A032140_20210817T152909
L2A_T27XVG_A032154_20210818T145938
L2A_T26XNM_A032154_20210818T145938
L2A_T27XVG_A032168_20210819T142935
L2A_T26XNM_A032168_20210819T142935
L2A_T27XVG_A023260_20210819T151803
L2A_T26XNM_A023260_20210819T151803
L2A_T27XVG_A023274_20210820T144750
L2A_T26XNM_A023274_20210820T144750
L2A_T27XVG_A032197_20210821T150913
L2A_T26XNM_A032197_20210821T150913
L2A_T27XVG_A032211_20210822T143925
L2A_T26XNM_A032211_20210822T143925
L2A_T27XVG_A023303_20210822T152807
L2A_T26XNM_A023303_20210822T152807
L2A_T27XVG_A023317_20210823T145754
L2A_T26XNM_A023317_20210823T145754
L2A_T27XVG_A023331_20210824T142741
L2A_T26XNM_A023331_20210824T142741
L2A_T27XVG_A032240_20210824T151911
L2A_T26XNM_A032240_20210824T151911
L2A_T27XVG_A032254_20210825T144920
L2A_T26XNM_A032254_20210825T144920
L2A_T27XVG_A023360_20210826T150906
L2A_T26XNM_A023360_20210826T150906
L2A_T27XVG_A023374_20210827T143745
L2A_T26XNM_A023374_20210827T143745
L2A_T27XVG_A032283_20210827T152908
L2A_T26XNM_A032283_20210827T152908
Add a Sentinel-2 image to the map#
The visible bands in Sentinel-2 are 'B2':blue, 'B3':green, 'B4':red.
So to show a “true color” RGB composite image on the map, we need to select these bands in the R-G-B order:
myImage = collection.first()
myImage_RGB = myImage.select('B4', 'B3', 'B2')
vis_params = {'min': 0.0, 'max': 10000, 'opacity': 1.0, 'gamma': 1.5}
Map.addLayer(myImage_RGB, vis_params, name='my image')
Map.addLayer(gtx_feature,{'color': 'red'},'ground track')
Map
This seems to have worked. But there’s clouds everywhere.
Calculate the along-track cloud probability#
We need a better approach to get anywhere here. To do this, we use not only the Sentinel-2 Surface Reflectance image collection, but also merge it with the Sentinel-2 cloud probability collection, which can be accessed under COPERNICUS/S2_CLOUD_PROBABILITY.
Let’s specify a function that adds the cloud probability band to each Sentinel-2 image and calcultes the mean cloud probability in the neighborhood of the ICESat-2 ground track, then map this function over our location/date filtered collection.
def get_sentinel2_cloud_collection(is2data, days_buffer=6, gt_buffer=100):
# create the area of interest for cloud likelihood assessment
ground_track_coordinates = list(zip(is2data.atl08.lon, is2data.atl08.lat))
ground_track_projection = 'EPSG:4326' # our data is lon/lat in degrees [https://epsg.io/4326]
gtx_feature = ee.Geometry.LineString(coords=ground_track_coordinates,
proj=ground_track_projection,
geodesic=True)
area_of_interest = gtx_feature.buffer(gt_buffer)
datetime_requested = datetime.strptime(is2data.date, '%Y-%m-%d')
start_date = (datetime_requested - timedelta(days=days_buffer)).strftime('%Y-%m-%dT%H:%M:%S')
end_date = (datetime_requested + timedelta(days=days_buffer)).strftime('%Y-%m-%dT%H:%M:%S')
print('Search for imagery from {start:s} to {end:s}.'.format(start=start_date, end=end_date))
# Import and filter S2 SR HARMONIZED
s2_sr_collection = (ee.ImageCollection('COPERNICUS/S2_SR_HARMONIZED')
.filterBounds(area_of_interest)
.filterDate(start_date, end_date))
# Import and filter s2cloudless.
s2_cloudless_collection = (ee.ImageCollection('COPERNICUS/S2_CLOUD_PROBABILITY')
.filterBounds(area_of_interest)
.filterDate(start_date, end_date))
# Join the filtered s2cloudless collection to the SR collection by the 'system:index' property.
cloud_collection = ee.ImageCollection(ee.Join.saveFirst('s2cloudless').apply(**{
'primary': s2_sr_collection, 'secondary': s2_cloudless_collection,
'condition': ee.Filter.equals(**{'leftField': 'system:index','rightField': 'system:index'})}))
cloud_collection = cloud_collection.map(lambda img: img.addBands(ee.Image(img.get('s2cloudless')).select('probability')))
def set_is2_cloudiness(img, aoi=area_of_interest):
cloudprob = img.select(['probability']).reduceRegion(reducer=ee.Reducer.mean(),
geometry=aoi,
bestEffort=True,
maxPixels=1e6)
return img.set('ground_track_cloud_prob', cloudprob.get('probability'))
return cloud_collection.map(set_is2_cloudiness)
Get this collection for our ICESat-2 data, and print all the granule IDs and associated cloudiness over the ground track.
collection = get_sentinel2_cloud_collection(is2data)
info = collection.getInfo()
for feature in info['features']:
print('%s --> along-track cloud probability: %5.1f %%' % (feature['properties']['GRANULE_ID'],
feature['properties']['ground_track_cloud_prob']))
Search for imagery from 2021-08-16T00:00:00 to 2021-08-28T00:00:00.
L2A_T26XNM_A023217_20210816T150915 --> along-track cloud probability: 89.7 %
L2A_T27XVG_A023217_20210816T150915 --> along-track cloud probability: 89.7 %
L2A_T26XNM_A023231_20210817T143747 --> along-track cloud probability: 4.5 %
L2A_T27XVG_A023231_20210817T143747 --> along-track cloud probability: 4.6 %
L2A_T26XNM_A032140_20210817T152909 --> along-track cloud probability: 12.1 %
L2A_T27XVG_A032140_20210817T152909 --> along-track cloud probability: 12.1 %
L2A_T26XNM_A032154_20210818T145938 --> along-track cloud probability: 10.3 %
L2A_T27XVG_A032154_20210818T145938 --> along-track cloud probability: 10.1 %
L2A_T26XNM_A032168_20210819T142935 --> along-track cloud probability: 12.9 %
L2A_T27XVG_A032168_20210819T142935 --> along-track cloud probability: 12.8 %
L2A_T26XNM_A023260_20210819T151803 --> along-track cloud probability: 9.8 %
L2A_T27XVG_A023260_20210819T151803 --> along-track cloud probability: 9.9 %
L2A_T26XNM_A023274_20210820T144750 --> along-track cloud probability: 1.3 %
L2A_T27XVG_A023274_20210820T144750 --> along-track cloud probability: 1.3 %
L2A_T26XNM_A032197_20210821T150913 --> along-track cloud probability: 20.2 %
L2A_T27XVG_A032197_20210821T150913 --> along-track cloud probability: 20.3 %
L2A_T26XNM_A032211_20210822T143925 --> along-track cloud probability: 14.8 %
L2A_T27XVG_A032211_20210822T143925 --> along-track cloud probability: 14.8 %
L2A_T26XNM_A023303_20210822T152807 --> along-track cloud probability: 15.7 %
L2A_T27XVG_A023303_20210822T152807 --> along-track cloud probability: 15.8 %
L2A_T26XNM_A023317_20210823T145754 --> along-track cloud probability: 5.6 %
L2A_T27XVG_A023317_20210823T145754 --> along-track cloud probability: 5.5 %
L2A_T26XNM_A023331_20210824T142741 --> along-track cloud probability: 8.0 %
L2A_T27XVG_A023331_20210824T142741 --> along-track cloud probability: 7.9 %
L2A_T26XNM_A032240_20210824T151911 --> along-track cloud probability: 7.5 %
L2A_T27XVG_A032240_20210824T151911 --> along-track cloud probability: 7.5 %
L2A_T26XNM_A032254_20210825T144920 --> along-track cloud probability: 7.7 %
L2A_T27XVG_A032254_20210825T144920 --> along-track cloud probability: 7.8 %
L2A_T26XNM_A023360_20210826T150906 --> along-track cloud probability: 15.8 %
L2A_T27XVG_A023360_20210826T150906 --> along-track cloud probability: 15.8 %
L2A_T26XNM_A023374_20210827T143745 --> along-track cloud probability: 10.3 %
L2A_T27XVG_A023374_20210827T143745 --> along-track cloud probability: 10.1 %
L2A_T26XNM_A032283_20210827T152908 --> along-track cloud probability: 9.8 %
L2A_T27XVG_A032283_20210827T152908 --> along-track cloud probability: 9.8 %
Filter cloudy images#
We specify a certain cloud probability threshold, and then only keep the images that fall below it. Here we are choosing a quite aggressive value of maximum 5% cloud probability…
# filter by maximum allowable cloud probability (in percent)
MAX_CLOUD_PROB_ALONG_TRACK = 5
cloudfree_collection = collection.filter(ee.Filter.lt('ground_track_cloud_prob', MAX_CLOUD_PROB_ALONG_TRACK))
print('There are %i cloud-free scenes.' % cloudfree_collection.size().getInfo())
There are 4 cloud-free scenes.
Sort the collection by time difference from the ICESat-2 overpass#
Using the image property 'system:time_start' we can calculate the time difference from the ICESat-2 overpass and set it as a property. This allows us to sort the collection by it and to make sure that the first image in the collection is the closest-in-time to ICESat-2 image that is also cloud-free.
# get the time difference between ICESat-2 overpass and Sentinel-2 acquisitions, set as image property
is2time = is2data.date + 'T12:00:00'
def set_time_difference(img, is2time=is2time):
timediff = ee.Date(is2time).difference(img.get('system:time_start'), 'second').abs()
return img.set('timediff', timediff)
cloudfree_collection = cloudfree_collection.map(set_time_difference).sort('timediff')
Print some stats for the final collection to make sure everything looks alright.
info = cloudfree_collection.getInfo()
for feature in info['features']:
s2datetime = datetime.fromtimestamp(feature['properties']['system:time_start']/1e3)
is2datetime = datetime.strptime(is2time, '%Y-%m-%dT%H:%M:%S')
timediff = s2datetime - is2datetime
timediff -= timedelta(microseconds=timediff.microseconds)
diffsign = 'before' if timediff < timedelta(0) else 'after'
print('%s --> along-track cloud probability: %5.1f %%, %s %7s ICESat-2' % (feature['properties']['GRANULE_ID'],
feature['properties']['ground_track_cloud_prob'],np.abs(timediff), diffsign))
L2A_T26XNM_A023274_20210820T144750 --> along-track cloud probability: 1.3 %, 1 day, 21:09:34 before ICESat-2
L2A_T27XVG_A023274_20210820T144750 --> along-track cloud probability: 1.3 %, 1 day, 21:09:38 before ICESat-2
L2A_T26XNM_A023231_20210817T143747 --> along-track cloud probability: 4.5 %, 4 days, 21:19:32 before ICESat-2
L2A_T27XVG_A023231_20210817T143747 --> along-track cloud probability: 4.6 %, 4 days, 21:19:36 before ICESat-2
Show the final image and ground track on the map#
first_image_rgb = cloudfree_collection.first().select('B4', 'B3', 'B2')
Map = geemap.Map(layout=Layout(width='70%', max_height='450px'))
Map.add_basemap('SATELLITE')
Map.addLayer(first_image_rgb, vis_params, name='my image')
Map.addLayer(gtx_feature,{'color': 'red'},'ground track')
Map.centerObject(gtx_feature, zoom=12)
Map
Download images from Earth Engine#
We can use .getDownloadUrl() on an Earth Engine image.
It asks for a scale, which is just the pixel size in meters (10 m for Sentinel-2 visible bands). It also asks for the region we would like to export; here we use a .buffer around the center.
(Note: This function can only be effectively used for small download jobs because there is a request size limit. Here, we only download a small region around the ground track, and convert the image to an 8-bit RGB composite to keep file size low. For larger jobs you should use Export.image.toDrive)
# create a region around the ground track over which to download data
point_of_interest = ee.Geometry.Point(center_lon, center_lat)
buffer_around_center_meters = ground_track_length*0.52
region_of_interest = point_of_interest.buffer(buffer_around_center_meters)
# make the image 8-bit RGB
s2rgb = first_image_rgb.unitScale(ee.Number(0), ee.Number(10000)).clamp(0.0, 1.0).multiply(255.0).uint8()
# get the download URL
downloadURL = s2rgb.getDownloadUrl({'name': 'mySatelliteImage',
'crs': s2rgb.projection().crs(),
'scale': 10,
'region': region_of_interest,
'filePerBand': False,
'format': 'GEO_TIFF'})
downloadURL
'https://earthengine.googleapis.com/v1/projects/earthengine-legacy/thumbnails/5748d80a6b75e56a6f4cbc6329613330-e48a892424305a54a523e0af626197f4:getPixels'
We can save the content of the download URL with the requests library.
response = requests.get(downloadURL)
filename = 'my-satellite-image.tif'
with open(filename, 'wb') as f:
f.write(response.content)
print('Downloaded %s' % filename)
Downloaded my-satellite-image.tif
Open a GeoTIFF in rasterio#
Now that we have saved the file, we can open it locally with the rasterio library.
myImage = rio.open(filename)
myImage
<open DatasetReader name='my-satellite-image.tif' mode='r'>
Plot a GeoTIFF in Matplotlib#
Now we can easily plot the image in a matplotlib figure, just using the rasterio.plot() module.
fig, ax = plt.subplots(figsize=[4,4])
rioplot.show(myImage, ax=ax);
transform the ground track into the image CRS#
Because our plot is now in the Antarctic Polar Stereographic Coordrinate Reference System, we need to project the coordinates of the ground track from lon/lat values. The rasterio.warp.transform function has us covered. From then on, it’s just plotting in Matplotlib.
gtx_x, gtx_y = warp.transform(src_crs='epsg:4326', dst_crs=myImage.crs, xs=is2data.atl08.lon, ys=is2data.atl08.lat)
ax.plot(gtx_x, gtx_y, color='red', linestyle='-')
ax.axis('off')
(570580.0, 575140.0, 8608010.0, 8612600.0)
Putting it all together#
The code above is found more concisely in one more method:
dataCollector.visualize_sentinel2()
This one has some default parameters, which you can change when running it:
- max_cloud_prob = 20
- days_buffer = 10
- gamma_value = 1.8
- title = 'ICESat-2 data'
- imagery_filename = 'my-satellite-image.tif'
- plot_filename = 'my-plot.jpg'
We can now do everything we did in this tutorial in just three lines!
url = 'http://openaltimetry.org/data/api/icesat2/atl03?date=2021-08-22&minx=-23.989069717620517&miny=77.53261687779506&maxx=-23.950348700496775&maxy=77.57222464044443&trackId=909&beamName=gt1r&beamName=gt1l&outputFormat=json'
is2data = dataCollector(oaurl=url,beam='gt1r')
fig = is2data.visualize_sentinel2(max_cloud_prob=5, title='some ICESat-2 data with a Sentinel-2 image')
--> Getting data from OpenAltimetry.
---> requesting ATL03 data... 6314 data points.
---> requesting ATL06 data... 198 data points.
---> requesting ATL07 data... No data.
---> requesting ATL08 data... 26 data points.
---> requesting ATL10 data... No data.
---> requesting ATL12 data... No data.
---> requesting ATL13 data... No data.
The ground track is 4.4 km long.
Looking for Sentinel-2 images from 2021-08-12T12:00:00 to 2021-09-01T12:00:00 --> there are 4 cloud-free images.
Looking for Sentinel-2 images from 2021-08-02T12:00:00 to 2021-09-11T12:00:00 --> there are 8 cloud-free images.
Looking for Sentinel-2 images from 2021-07-23T12:00:00 to 2021-09-21T12:00:00 --> there are 24 cloud-free images.
--> Closest cloud-free Sentinel-2 image to ICESat:
- product_id: S2B_MSIL2A_20210820T144749_N0301_R082_T26XNM_20210820T173503
- time difference: 2 days before ICESat-2
- mean along-track cloud probability: 1.3
--> Downloaded the 8-bit RGB image as my-satellite-image.tif.
--> Saved plot as my-plot.jpg.
And now we can also easily run this on some totally different data!
url = 'http://openaltimetry.org/data/api/icesat2/atl12?date=2020-12-15&minx=-77.858681&miny=25.728091&maxx=-77.831461&maxy=25.832559&trackId=1254&beamName=gt1r&beamName=gt1l&outputFormat=json'
mydata = dataCollector(oaurl=url, beam='gt1r')
fig = mydata.visualize_sentinel2(max_cloud_prob=20,
title='Nearshore Bathymetry ICESat-2 Data',
imagery_filename='my-other-satellite-image.tif',
plot_filename='nearshore-bathymetry.jpg')
--> Getting data from OpenAltimetry.
---> requesting ATL03 data... 34237 data points.
---> requesting ATL06 data... No data.
---> requesting ATL07 data... No data.
---> requesting ATL08 data... 114 data points.
---> requesting ATL10 data... No data.
---> requesting ATL12 data... 2 data points.
---> requesting ATL13 data... 317 data points.
The ground track is 11.6 km long.
Looking for Sentinel-2 images from 2020-12-05T12:00:00 to 2020-12-25T12:00:00 --> there are 4 cloud-free images.
Looking for Sentinel-2 images from 2020-11-25T12:00:00 to 2021-01-04T12:00:00 --> there are 10 cloud-free images.
--> Closest cloud-free Sentinel-2 image to ICESat:
- product_id: S2A_MSIL2A_20201213T155521_N0214_R011_T17RRJ_20201213T195958
- time difference: 2 days before ICESat-2
- mean along-track cloud probability: 5.7
--> Downloaded the 8-bit RGB image as my-other-satellite-image.tif.
--> Saved plot as nearshore-bathymetry.jpg.
Exercise#
Find some data from OpenAltimetry, and plot it with a cloud-free satellite image.
Look for small-scale features - say a few hundred meters to 20 kilometers along-track. (Hint: OpenAltimetry has a scale bar.)
Bonus points for features where concurrent imagery and the ATL03 photon cloud may give us some information that we would not get from higher-level products alone!
Note: There is no Sentinel-2 coverage at higher latitudes than ~84°, and there will be no concurrent imagery during polar night.
If you don’t know where to start with OpenAltimetry, you can look here.
##### YOUR CODE GOES HERE - uncomment and edit it!
# url = 'http://???.org/??'
# gtx = 'gt??'
# is2data = dataCollector(oaurl=url, beam=gtx)
# fig = is2data.visualize_sentinel2(title='<your figure title goes here>',
# imagery_filename='your-satellite-image.tif',
# plot_filename='your-plot.jpg')
Summary#
🎉 Congratulations! You’ve completed this tutorial and have seen how we can put ICESat-2 photon-level data into context using Google Earth Engine and the OpenAltimetry API.
You can explore a few more use cases for this code (and possible solutions for Exercise 2) in this notebook.
References#
To further explore the topics of this tutorial see the following detailed documentation:
Bonus Material: Code for Extracting Along-Track Surface Reflectance Values#
Based on javascript code in this example.
# a function for buffering ICESat-2 along-track depth measurement locations by a footprint radius
def bufferPoints(radius, bounds=False):
def buffer(pt):
pt = ee.Feature(pt)
return pt.buffer(radius).bounds() if bounds else pt.buffer(radius)
return buffer
# a function for extracting Sentinel-2 band data for ICESat-2 along-track depth measurement locations
def zonalStats(ic, fc, reducer=ee.Reducer.mean(), scale=None, crs=None, bands=None, bandsRename=None,
imgProps=None, imgPropsRename=None, datetimeName='datetime', datetimeFormat='YYYY-MM-dd HH:mm:ss'):
# Set default parameters based on an image representative.
imgRep = ic.first()
nonSystemImgProps = ee.Feature(None).copyProperties(imgRep).propertyNames()
if not bands:
bands = imgRep.bandNames()
if not bandsRename:
bandsRename = bands
if not imgProps:
imgProps = nonSystemImgProps
if not imgPropsRename:
imgPropsRename = imgProps
# Map the reduceRegions function over the image collection.
results = ic.map(lambda img:
img.select(bands, bandsRename)
.set(datetimeName, img.date().format(datetimeFormat))
.set('timestamp', img.get('system:time_start'))
.reduceRegions(collection=fc.filterBounds(img.geometry()),reducer=reducer,scale=scale,crs=crs)
.map(lambda f: f.set(img.toDictionary(imgProps).rename(imgProps,imgPropsRename)))
).flatten().filter(ee.Filter.notNull(bandsRename))
return results
ground_track_buffer = 7.5 # radius in meters, for a 15-m diameter footprint
ground_track_coordinates = list(zip(is2data.gt.lon, is2data.gt.lat))
gtx_feature = ee.Geometry.LineString(coords=ground_track_coordinates, proj='EPSG:4326', geodesic=True)
aoi = gtx_feature.buffer(ground_track_buffer)
# clip to the ground track for along-track querying
img_gt = cloudfree_collection.first().clip(aoi)
# get the footprint-buffered query points from the lake object's depth_data dataframe
pts = ee.FeatureCollection([
ee.Feature(ee.Geometry.Point([r.lon, r.lat]), {'plot_id': i}) for i,r in is2data.gt.iterrows()
])
ptsS2 = pts.map(bufferPoints(ground_track_buffer))
# query the Sentinel-2 bands at the ground track locations where lake depths are posted
thiscoll = ee.ImageCollection([img_gt])
bandNames = ['B1', 'B2', 'B3', 'B4', 'B5', 'B6', 'B7', 'B8', 'B8A', 'B9', 'B11', 'B12', 'probability']
bandIdxs = list(range(len(bandNames)-1)) + [23]
ptsData = zonalStats(thiscoll, ptsS2, bands=bandIdxs, bandsRename=bandNames, imgProps=['PRODUCT_ID'], scale=5)
# get the data to the client side and merge into the lake depth data frame
features_select = ['plot_id'] + bandNames
results = ptsData.select(features_select, retainGeometry=False).getInfo()['features']
data = [x["properties"] for x in results]
dfS2 = pd.DataFrame(data).set_index('plot_id')
df_bands = is2data.gt.join(dfS2, how='left')
# calculate the Normalized Difference Water Index (NDWI)
df_bands['ndwi'] = (df_bands.B2-df_bands.B4) / (df_bands.B2+df_bands.B4)
df_bands
| index | lat | lon | xatc | B1 | B11 | B12 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | B8A | B9 | probability | ndwi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 499 | 77.572104 | -23.954022 | 0.000000 | 7542.079621 | 164.685412 | 112.618040 | 7679.957684 | 7304.628062 | 6419.345212 | 6360.185412 | 5687.488864 | 4959.208797 | 4554.002227 | 4227.100223 | 4271.315702 | 0.000000 | 0.089410 |
| 1 | 498 | 77.572025 | -23.954088 | 8.906222 | 7519.000000 | 164.866406 | 112.482392 | 7673.486864 | 7283.530464 | 6385.437675 | 6358.749581 | 5691.365567 | 4957.435998 | 4589.643376 | 4232.547233 | 4267.000000 | 0.000000 | 0.091618 |
| 2 | 497 | 77.571946 | -23.954154 | 17.812443 | 7519.000000 | 164.214807 | 110.955132 | 7667.717330 | 7348.897364 | 6504.787437 | 6403.295008 | 5750.389792 | 4981.712283 | 4688.150308 | 4231.484016 | 4267.000000 | 0.000000 | 0.082055 |
| 3 | 496 | 77.571867 | -23.954220 | 26.718665 | 7519.000000 | 163.100390 | 110.193530 | 7614.340212 | 7219.821528 | 6464.990519 | 6421.064696 | 5779.469604 | 4995.268823 | 4696.385945 | 4231.175683 | 4267.000000 | 0.000000 | 0.081634 |
| 4 | 495 | 77.571789 | -23.954286 | 35.624887 | 7519.000000 | 163.584213 | 104.806559 | 7571.846581 | 7152.758199 | 6384.584769 | 6316.176209 | 5657.226237 | 4909.948860 | 4619.942190 | 4142.191217 | 4267.000000 | 0.000000 | 0.085069 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 495 | 4 | 77.533083 | -23.986586 | 4408.579718 | 8041.000000 | 201.959686 | 124.979843 | 8068.022396 | 7621.547592 | 6661.511758 | 6541.088466 | 5891.798432 | 5152.294513 | 4894.360582 | 4448.604703 | 4480.000000 | 0.386338 | 0.095489 |
| 496 | 3 | 77.533004 | -23.986653 | 4417.485940 | 8041.000000 | 200.616331 | 124.308166 | 8119.082774 | 7650.724832 | 6645.447427 | 6577.359060 | 5885.081655 | 5128.785794 | 4903.615213 | 4468.755034 | 4480.000000 | 0.084452 | 0.099809 |
| 497 | 2 | 77.532925 | -23.986717 | 4426.392162 | 8040.609131 | 199.819599 | 123.969933 | 8081.496659 | 7619.690423 | 6637.523385 | 6592.406459 | 5883.623608 | 5117.819599 | 4823.741648 | 4475.865256 | 4477.143653 | 0.152004 | 0.098103 |
| 498 | 1 | 77.532846 | -23.986783 | 4435.298383 | 8039.849750 | 189.611018 | 127.637173 | 8041.181970 | 7589.139677 | 6624.518642 | 6584.066778 | 5910.378408 | 5111.065109 | 4767.759599 | 4448.170840 | 4471.594324 | 0.152476 | 0.096597 |
| 499 | 0 | 77.532768 | -23.986849 | 4444.204605 | 8038.890128 | 180.460680 | 131.649191 | 8112.696040 | 7589.586168 | 6630.025655 | 6579.622978 | 5937.141104 | 5107.816509 | 4779.498048 | 4421.994980 | 4464.581707 | 0.485778 | 0.100570 |
500 rows × 18 columns
fig, axs = plt.subplots(figsize=[9,5], nrows=3, sharex=True)
ax = axs[0]
ax.scatter(is2data.atl03.lat, is2data.atl03.h, s=1, c='k', label='ATL03')
ax.set_ylim((741, 818))
ax.legend()
ax = axs[1]
ax.plot(df_bands.lat,df_bands.ndwi, c='b', label='NDWI')
ax.set_ylim((0, 1))
ax.legend()
ax = axs[2]
scale = 1e-4
ax.plot(df_bands.lat,df_bands.B4*scale, c='r', label='B4')
ax.plot(df_bands.lat,df_bands.B3*scale, c='g', label='B3')
ax.plot(df_bands.lat,df_bands.B2*scale, c='b', label='B2')
ax.set_ylim((0, 1))
ax.legend()
ax.set_xlim((is2data.gt.lat.min(), is2data.gt.lat.max()))
ax.set_xlabel('latitude')
Text(0.5, 0, 'latitude')

